4.6 Using Headerlets
Introduction
A headerlet is a compact FITS file representing a single WCS solution for a single exposure that has been aligned to a particular image or astrometric catalog, complete with all distortion information. It could be utilized as a convenient WCS information package that a user could e-mail to a collaborator working on the same image, so it can be incorporated into the collaborator’s copy of the image. This avoids the need to send that entire image containing the new WCS to the collaborator.
Headerlets serve two primary purposes:
- They contain a summary of all WCS information for a single exposure.
- They can be used to update the WCS information in additional copies of the same image.
The default suffix for headerlet files, as used by all DrizzlePac tasks, is hlet.fits. For example, the name of a headerlet written out by tweakreg for the WFC3 image iabf01ckq_flt.fits for the WCS solution named TWEAK would be named iabf01ckq_tweak_hlet.fits.
Why Headerlets are Useful
A lot of effort can be expended in determining the alignment between an image and an astrometric field or another image. It requires some expertise with source-finding algorithms, computing offset fit solutions, and a knowledge of FITS WCS standards. That expertise can now be embodied in headerlets created using the tweakreg task, which was written to simplify image alignment and provide updated WCS information for aligned images. This information, stored in the headerlets, can then be easily applied to the same images being studied by other users, saving them the work needed to align the images to the same reference image or catalog.
Types of Information Contained in Headerlets
AstroDrizzle requires that all WCS and distortion information be present in the input images in order to align them. This can include multiple components, some that are time-dependent, and others that are exposure- or orbit-dependent. The updatewcs task, used by AstroDrizzle in the pipeline (and also available to users), reads in base models from the reference files; it computes components such as time- and orbit-dependent WCS information that are unique to an exposure, and populates the relevant WCS keywords in the image header. For example, ACS and WFC3 data have a velocity aberration keyword which reports the specific aberration for that exposure. That information gets applied to the distortion model as an additional plate scale correction. As a result, the distortion solution for one image will never be applicable to any other exposure, even if they are taken back-to-back.
When aligning a set of images from different visits, image WCS offsets between visits due to guide star coordinate errors need to be removed. This involves revising the image WCSs such that they align with one image from the set that acts as the reference image. Another option is to align the image WCSs to an external astrometric catalog. Therefore, updating the WCS, as done by tweakreg, accounts not only for the full distortion model (as mentioned in the previous paragraph), it can also correct WCS offsets due to different guide stars.
WCS Information in Archival Images
For active instruments1 (specifically, ACS and WFC3), when a user submits a data request to the HST Archive, images are processed with the latest image calibrations, including the latest available distortion models, by the On-The-Fly Reprocessing (OTFR) system before being delivered to the user. Calibrated image files, that have the suffix flt.fits, (or flc.fits for CTE-corrected ACS images), serve as inputs to AstroDrizzle where distortions are corrected and the images are drizzle-combined into a single drizzled image (with suffix drz.fits, or drc.fits from flc.fits input files).
WCS information in flt.fits images gets updated when AstroDrizzle is run in the pipeline. The full distortion model, including the full polynomial solution from the IDCTAB reference file, and all the corrections formerly combined into the DGEOFILE reference image, are now stored directly in the images in the form of keyword values and FITS extensions. This report contains a description of the conventions for coordinate transformations used to describe all these components in a FITS file.
As described in Section 3.3.2, image headers now contain the following keywords and image extensions to fully describe the WCS with distortion corrections.
- Linear WCS (including CD matrix) keywords: CRPIX*, CRVAL*, CD*, and CTYPE*
- SIP coefficients keywords: A_p_q and B_p_q2, A_ORDER, B_ORDER, OCX10, OCX11, OCY10, and OCY11
- 3Residual distortion corrections reference file: if the NPOLFILE header keyword specifies the name of a residual distortions reference file for an image, CPDIS* and DP* keywords point to image extensions of type WCSDVARR (Paper IV convention)
- 7WCSDVARR extensions: two image extensions per chip are created, each with look-up tables containing the corrections from the NPOLFILE (non-polynomial) reference file, where each extension corresponds to an image axis (X correction or Y correction)
- 7Column correction reference file: if the D2IMFILE header keyword specifies a reference file for column width corrections, the D2IMEXT and D2IMERR keywords point to a file extension of type D2IMVARR.
- 7D2IMVARR extension: an extension with a look-up table containing chip column corrections from the D2IMFILE reference file.
Each science header will have its own set of these keywords, as well as extensions that will be kept together as part of the full WCS solution.
Only one set of distortion coefficients and look-up tables, stored in the SIP header keywords and from the NPOLFILE reference file, respectively, can be included in the science header. This single distortion model is used by all alternate WCS’s in the image header that are stored as sets of WCS keywords using the FITS Paper I Multiple WCS Standard.
Storing Multiple WCSs in an image
When making updates to any WCS solution, the software will, by default, save previous WCS solutions in case it is needed for later use. This results in multiple alternate WCS solutions, based on the same distortion model, being present in the image header at one time. Therefore, each image has the potential to align to several different astrometric solutions, depending on which alternate WCS solution the user wants to select. One potential reason for having multiple headerlets in an image would be to provide WCSs that are aligned to different astrometric catalogs. That would allow a user to pick a WCS that matches a catalog that he or she wishes to use. These layers of multiple WCS solutions can be somewhat complex. Code in the STWCS package, used by DrizzlePac tasks, was designed to simplify the management of these different WCSs, making them more easily accessible via headerlets and other drizzlepac task parameters.
Even though there are a large number of extensions appended to this FITS file, the sum total of all of these new extensions comes to approximately 100kB for ACS/WFC images, making them a space efficient means of managing all of the distortion and WCS information.
WCSCORR Image Extension for Storing Multiple WCSs
The code block in Section 4.3 shows an entry that has yet to be mentioned: WCSCORR. This is a FITS extension in the form of a binary table that was first created when running tweakreg with updatehdr=True and updated every time tweakreg is run with updatehdr=True to keep a record of all updates made to WCS keywords. Each row of the WCSCORR table corresponds to a single update to a single chip's set of WCS keywords. Each set of keywords defines what needs to be included in a headerlet when it gets written as a WCS solution for a single exposure.
The columns of the table include, at minimum, these keywords associated with a specific WCS:
| Unique descriptive ID for this particular solution |
| Chip to which this solution applies |
| FITS Paper I Alternate WCS ID |
| Label for headerlet solution |
| Name of polynomial distortion solution |
| Name of non-polynomial distortion |
| Name of detector-to-image distortion |
| New value of CRVAL1 |
| New value of CRVAL2 |
| New value of CRPIX1 |
| New value of CRPIX2 |
| New value of CD1_1 |
| New value of CD1_2 |
| New value of CD2_1 |
| New value of CD2_2 |
| Name of coordinate for axis 1 |
| Name of coordinate for axis 2 |
| New value of ORIENTAT (derived from new CD) |
| New value of PA_V3 (required for updatewcs) |
| RMS in RA for fit (arcseconds) |
| RMS in Dec for fit (arcseconds) |
| Number of sources used in fit |
| Name of astrometric catalog used for fit |
| Description of fit or sources used in fit |
Syntax for Alternate WCSs
Details of how alternate WCSs get defined and used in FITS headers can be found online in FITS WCS Paper I. This capability is crucial to the success of the DrizzlePac software, as this standard allows multiple alignment solutions for an image to be specified in the image header at one time.
The default WCS solution is referred to as the primary WCS in documentation for DrizzlePac and STWCS tasks as it serves as the primary or default WCS transformation that is applied to the pixel positions.
Each alternate WCS solution is identified by a single letter, referred to in the standard and this documentation as a wcskey, with values from A to Z. The primary WCS, however, is assigned a wcskey of “ ” (a string comprised of a single blank). All keywords that make up each alternate WCS solution has a wcskey value appended to the end of the keyword name. For example, the alternate WCS keywords with the wcskey of “A” would have keywords CRVAL1A, CRVAL2A, CRPIX1A, CRPIX2A, and so on.
In addition, each WCS solution, including the primary WCS, gets labeled using the WCSNAME* keyword with the WCS’s key appended to the end of the keyword. For example, the alternate WCS associated with the wcskey of “A” would have the keyword named WCSNAMEA.
The wcskey of “O” has been reserved by DrizzlePac and STWCS packages for use in archiving the original WCS solution generated in the Archive. It will always be WCSNAMEO=“OPUS”. The software makes it nearly impossible to overwrite it since WCSNAMEO provides a means of recovering the original WCS solution in case updates to the images’s WCS keywords introduces errors that can only be corrected by starting from scratch. The updatewcs task uses the alternate “0” WCS to recompute the original WCS solution assuming the distortion reference files can still be accessed, as it assumes that none of the WCS or distortion information in the headers are accurate.
Headerlet Structure
Headerlets are generated using the headerlet tasks in the stwcs.wcsutil.headerlet module (more about this later in the section) or by running the tweakreg task with updatehdr=True. (Headerlets may also be created by other FITS standard software not covered in this document, as long as they follow established headeret definitions.)
When appended to a science FITS image, the headerlet is attached as a FITS extension with a non-standard extension type. This non-standard extension type will not cause problems with FITS readers, however, when using PyFITS, it will return the headerlet’s full FITS format with access to all the headerlet’s extensions4 when the STWCS package has been installed. For details on how to access a headerlet’s information when it is attached to an image, see “Accessing the Headerlet Extension using PyFITS”.
On the other hand, the STWCS package used by DrizzlePac tasks can use a headerlet to define a WCS for an image to perform coordinate transformations (complete with distortion correction) without even requiring the original science image and all its data.
Headerlet Format
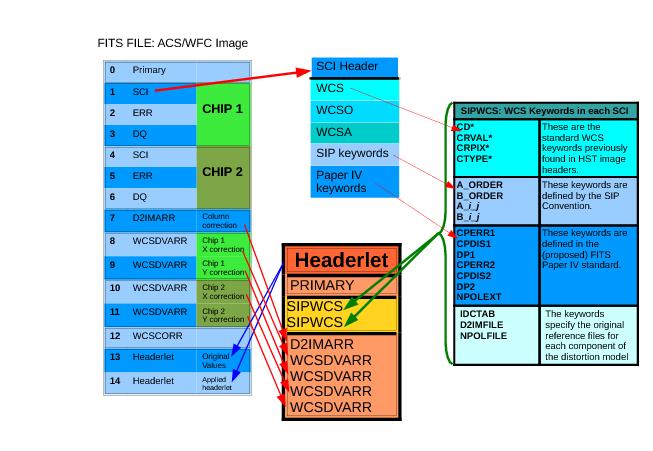
A headerlet is a multi-extension FITS file containing a single full WCS solution for a single exposure. The full set of extensions in a headerlet is determined by the distortion model used in the image, as well as the number of chips read out for each image. A PyFITS listing of extensions in a headerlet, created from an ACS/WFC image, is shown in "Accessing the Headerlet Extension using PyFITS". ACS images rely on non-polynomial (NPOL) and detector column width (D2IM) corrections in the form of look-up table extensions; these get included as extensions to the headerlet in order to maintain a complete set of distortion model information. The full set of keywords included in the headerlet and from where they are derived can be seen in Figure 4.1 below.
Note that a headerlet derived from a full-frame WFC3/UVIS image would only contain a PRIMARY header and two SIPWCS extensions (one for each SCI extension) as WFC3/UVIS does not currently use NPOLFILE or D2IMFILE reference files as part of their distortion model.
Headerlet’s PRIMARY Header
The PRIMARY header of a headerlet will only contain those keywords necessary for identifying the science exposure name, distortion model information, and title for the WCS solution. The primary header must have four required keywords:
HDRNAME, a unique name for the headerletDESTIM, target image filename (the ROOTNAME keyword of the original archive filename)STWCSVER, version of stwcs used to create the WCS of the original imagePYWCSVER, version of PyWCS used to create the WCS of the original image
These keywords are used for determining whether a headerlet can be applied to a given exposure and how it needs to be applied. Additional keywords provide more information about the solution itself, how it was derived, and by whom, through use of the following keywords:
AUTHOR, name of person who created the headerletDESCRIP, short description of the headerlet solutionRMS_RA, RMS in R.A. at the reference pixel of the WCS stored in the headerlet solution, if updated from the Archive’s default WCSRMS_DEC, RMS in Dec. at the reference pixel of the WCS stored in the headerlet solution, if updated from the Archive’s default WCSNMATCH, number of sources used in the new solution fit, if updated from the Archive’s default WCSCATALOG, astrometric catalog used for headerlet solutionCOMMENT, long description of how the headerlet solution was derived, if updated from Archive’s default WCS
These keywords allow the headerlet to retain enough information about how the new solution was generated so that a user could determine if it can be applied to his or her copy of the image.
SIPWCS: A New WCS FITS Extension
All WCS-related keywords from the image’s SCI headers, including all keywords referring to NPOL and D2IM extensions, are used to create SIPWCS headerlet extensions. The number of SIPWCS extensions directly correlate to the number of SCI extensions in the original science image. For instance, an ACS/WFC or WFC3/UVS image would have [SIPWCS,1] derived from [SCI,1] and [SIPWCS,2] derived from [SCI,2].
Keywords in SIPWCS extensions can then be used to overwrite SCI header keywords when a headerlet gets used to update a science image. The SIPWCS extension therefore serves not only as a record of the specific WCS solution derived for an image, but also as the source of values for replacing the image SCI headers WCS solution when desired. Keywords recording alignment information in the SIPWCS headesr also provide a record of how much of an offset there is between this solution and the default OPUS-generated solution.
Working with Headerlets
A set of tasks in stwcs.wcsutil.headerlet have been developed to support interactive management of headerlets, either creating, applying, or even deleting headerlets from a given image.
| apply a headerlet to a file |
| save a WCS solution as a headerlet extension and write it out as a headerlet FITS file |
| attach a headerlet as an extension to a file |
| delete a headerlet extension from a file |
| write out a headerlet extension as a separate FITS file |
| print a summary of all headerlet extensions in a file |
| replace current WCS solution with the WCS solution from a headerlet extension |
| save a WCS solution as a separate headerlet FITS file |
All these tasks are part of the STWCS package distributed along with the DrizzlePac package in the STScI_Python public release. This section describes how to use the most commonly-needed of these tasks to create headerlet FITS files and then apply them to science images. The full descriptions of all the remaining tasks goes beyond the scope of this handbook at this time (but please check the DrizzlePac website for updates).
Creating a Headerlet
The tweakreg task 'create_headerlet' can generate headerlets for users when updating the input images with the fit that aligns those images to a reference image or catalog. Interactively, a headerlet can be created from any image using the 'write_headerlet' task that will create a headerlet, write out the headerlet to a separate headerlet FITS file, and then, optionally, attach it as an extension to a science image (if it has not already been saved as an extension). The parameters for this task are:
Parameter | Default | Description |
filename | Name(s) of science file(s) from which headerlets will be created and written.
| |
hdrname | Unique name for the headerlet, stored in the HDRNAME keyword, that will serve as the label for this headerlet. | |
output | None | Filename, or rootname, of the output headerlet FITS file to be written out by this task. |
sciext | SCI | Name (EXTNAME) of the extension that contains the WCS to be saved. |
wcsname | None | Name of the WCS to be used to create the headerlet. It will return without doing anything when a blank string is given as input. |
wcskey | None | Alternate WCS key for the WCS used to create the headerlet: values from A to Z, or “ ” (blank string), or PRIMARY. If a blank string or PRIMARY are specified, then it will create the headerlet from the primary WCS. |
destim | None | The value of this parameter gets written out as the DESTIM keyword in the headerlet to denote that this headerlet can only be applied to that specific image. If set to None, the ROOTNAME keyword value from the science image header is used. |
sipname | None | Name of unique file which should be read to obtain the polynomial distortion coefficients. For a value of None, the code looks for the keyword SIPNAME in the science header.
|
npolfile | None | Name of a unique file where non-polynomial distortions are stored. If the value None is given, the value gets determined by the code using:
|
d2imfile | None | Name of a unique file where the detector to image correction was stored. If a value of None is given, the code get the value using:
|
author | None | Name of user who created the headerlet, used as the value for the AUTHOR keyword in the headerlet’s PRIMARY header. |
descrip | None | Short description of the solution provided by the headerlet, added as the value to the DESCRIP keyword in the headerlet’s PRIMARY header. |
history | None | Long (possibly multi-line) description of the solution provided by the headerlet. These comments will be added as HISTORY cards to the headerlet’s PRIMARY header. If a filename is specified, it will format and attach all text from that file as the history. |
attach | True | Specify whether or not to attach this headerlet as a new extension. It will verify that no other headerlet extension has been created with the same hdrname value. |
clobber | False | If the output headerlet file already exists, this parameter specifies whether or not to overwrite that file |
logging | False | This parameter enables logging of the operations to a file. |
Basically, a headerlet can be created from a science image header and recorded with user-specified history and descriptions. This task can also be called directly using Python with the syntax:
>>> from stwcs.wcsutil import headerlet >>> headerlet.write_headerlet(filename, hdrname, output=None, sciext='SCI', wcsname=None, wcskey=None, destim=None, sipname=None, npolfile=None, d2imfile=None, author=None, descrip=None, history=None, nmatch=None, catalog=None, attach=True, clobber=False, logging=False)
All the parameters used in the Python call are identical to those described for interactive use through the TEAL GUI.
Applying a Headerlet
Updating an image retrieved from the HST Archive with a headerlet, for example, requires the use of the task 'apply_headerlet'. The full set of parameters used to run this task are:
Parameter | Default | Description |
|---|---|---|
filename | File name of science observation whose WCS solution will be updated by the headerlet. | |
hdrlet | The filename of the headerlet to be applied to the science image. | |
attach | True | If set to True (default), it will append the headerlet to the FITS science file as a new extension after updating the science header with the WCS solution in the headerlet. |
primary | True | This parameter specifies whether or not to replace the PRIMARY WCS with the WCS from headerlet. |
archive | True | If set to True (default), then before updating the image’s WCS, it will create a headerlet from the WCS solution already present in the science image header and add it as an extension to the science file. This allows the previous solutions to be backed up in the file in a way that allows the user to restore it as needed. |
force | False | If set to True, this will cause the headerlet to replace the current PRIMARY WCS even if it has a different distortion model. Applying a headerlet with one distortion model to an image which has a different distortion model can sometimes introduce errors in the coordinate transformations. |
wcskey | Key value (A to Z, except O) to be used when writing the headerlet’s WCS solution to the science header as an alternate WCS. If None, the next available key will be used. | |
wcsname | Name to be assigned to the headerlet’s WCS solution when writing it out to the science header as an alternate WCS. By default, it will use the WCSNAME keyword value from the headerlet itself. Headerlets require the use of the WCSNAME keyword, but this allows the user to change it as desired when applying it to a science file. | |
logging | False | This parameter enables logging of the tasks operations to a file. |
The apply_headerlet task does not simply drop a new WCS solution into an image, but takes care to remember the previous solution in case it needs to be restored. The full set of operations performed by this task (some optional) include:
- Create a headerlet from the original WCS solution in the science image (this step can be turned off).
- Copy all WCS information from the science image to an alternate WCS using FITS Paper I standards.
- Copy the WCS solution from the headerlet to the science observation.
- Update the WCSCORR table with the linear distortion corrections WCS keyword values and name of the SIP solution (based on the name of the reference files) for each SIPWCS extension from the headerlet, along with the keyword values from the PRIMARY header of the headerlet.
- Append the new headerlet to the science image as a single new extension (optional)
This process assumes that when an image header is updated with a headerlet, the new solution from the headerlet will become the prime WCS while providing the option to simply add the headerlets solution as an alternate WCS instead. The updated image can then be aligned to other images based on the original WCS or any headerlet WCS solution applied to that image.
The primary difficulty in applying a headerlet comes from insuring that the distortion model of the headerlet matches the distortion model described in the science image to be updated. Headerlets rely on the DISTNAME keyword to provide the description of the distortion models in the headerlet and in the science image. If the science image has a different distortion model than the one specified in the headerlet, the original distortion model from the science image gets moved into a headerlet and appended to the file as a new headerlet extension, then all the distortion information gets replaced with the new distortion information (keywords and distortion extensions) from the headerlet. The user can override this synchronization of models, if they absolutely feel the need, but this process insures that the WCS specified in the headerlet gets applied to the science image exactly as it was determined when creating the new solution for the headerlet. For most users, this will never be a concern unless they have the need to either redefine the distortion models based on their own calibrations or need to update data they retrieved with old distortion models with the latest calibrations.
This task can also be called directly using Python to apply the headerlet from a file to the PRIMARY WCS of a science file with this syntax:
>>> from stwcs.wcsutil import headerlet # only if it hasn’t already been called >>> headerlet.apply_headerlet_as_primary(filename,hdrlet,attach=True,archive=True, force=False,logging=False)
Alternatively, to apply the headerlet as an alternate WCS solution in the science image, the following syntax can be used:
>>>headerlet.apply_headerlet_as_alternate(filename,hdrlet,attach=True,wcskey=None, wcsname=None,logging=False)
All the parameters for both functions correspond to parameters already described for use with the TEAL GUI. Additional information about using Python syntax to run headerlet tasks are described in the next section.
Python Syntax of the Headerlet Tasks
The material covered in this section requires some experience working with Python.
This section describes the currently implemented Python syntax for working with headerlets supported by the stwcs.wcsutil.headerlet module. First, there’s a potentially confusing point that should be cleared up: a headerlet, as implemented, is simply a FITS file containing multiple extensions that contain all of the parameters necessary to reproduce the WCS solution in the science image from which it was created.
When a headerlet is applied to an image, a copy of the original WCS information is appended to the image’s HDU (Header Data Unit) list as a special extension HDU called a Headerlet HDU. A Headerlet HDU consists of a simple header describing the headerlet, and has as its data, the headerlet file itself, which may be compressed. A Headerlet HDU has an XTENSION value of HDRLET. Common FITS readers (such as fv) will see this extension as an image extension. PyFITS in conjunction with STWCS, on the other hand, allows the user to actually work with that data array of the headerlet HDU as if it were a separate headerlet file, complete with extensions, as described below. This makes it easy for users to verify and work with the contents of each headerlet HDU separately.
The Python syntax, on the other hand, works primarily with the Headerlet object; a Python class that contains all the WCS keyword information and distortion information, as well as built-in methods (akin to functions) for applying, writing out, or performing other operations on this object. The rest of this discussion will help document how to use the Headerlet object in Python tasks or scripts as needed.
Software Requirements
Working with headerlets only requires stwcs and all the tasks defined in it (such as those reported for use with TEAL when the commands import stwcs or reload(stwcs) are issued). This package comes as part of the standard STScI Python package and can be used independently of drizzlepac.
Getting Headerlet Function Names
The full set of functions available for use with headerlets can be found in the stwcs.wcsutil.headerlet module which contains functions that can be called to perform some predefined set of operations from start to end, while the Headerlet class provides basic functionality used by all the functions in this module and by the TEAL GUI.
The full list of functions can be obtained using Python’s own introspection functionality:
>>> from stwcs.wcsutil import headerlet >>> dir(headerlet)
Under ipython, a user can also use the help() function to get more specific help for any function listed in the module, such as:
>>> help(headerlet.create_headerlet)
Creating a Headerlet Object
The headerlet module in stwcs provides the createHeaderlet() function to simply create a Headerlet object from an input science image. This function actually gets called by the write_headerlet task and write_headerlet function from this package, both of which have already been described. The write_headerlet() function creates the headerlet from a file, then automatically writes out the headerlet to a file without actually returning the Headerlet object for continued use. As a result, the following describes how to create a Headerlet object for use in Python tasks or scripts.
The call for creating a Headerlet object uses the createHeaderlet() function with the following syntax:
>>> from stwcs.wcsutil import headerlet >>> hdrlet = headerlet.create_headerlet(filename,sciext=’SCI’,hdrname=None, destim=None,wcskey=" ",wcsname=None,sipname=None,npolfile=None,d2imfile=None, author=None,descrip=None,history=None,nmatch=None,catalog=None,logging=False, logmode=’w’)
All the parameters used by the create_headerlet() function correspond to already described parameters used by the write_headerlet task, with one exception. The logmode parameter allows the user to specify whether or not to overwrite (“w”) or append (“a”) to the log when writing out processing comments to a log file with logging=True.
An example of how to use this function to create a Headerlet object from the image ACS/WFC j94f05bgq_flt.fits and label the Headerlet with the name “VERSION1” would be:
>>> from stwcs.wcsutil import headerlet >>> hdrlet = headerlet.createHeaderlet(’j94f05bgq_flt.fits’, ’VERSION1’) >>> type(hdrlet) <class ’stwcs.wcsutil.headerlet.Headerlet’> >>> hdrlet.info() Filename: (No file associated with this HDUList) No. Name Type Cards Dimensions Format 0 PRIMARY PrimaryHDU 12 () 1 SIPWCS ImageHDU 111 () 2 SIPWCS ImageHDU 110 () 3 WCSDVARR ImageHDU 15 (65, 33) float32 4 WCSDVARR ImageHDU 15 (65, 33) float32 5 WCSDVARR ImageHDU 15 (65, 33) float32 6 WCSDVARR ImageHDU 15 (65, 33) float32 7 D2IMARR ImageHDU 12 (4096,) float32
Here, the Headerlet object is similar to a normal PyFITS HDUList object. createHeaderlet() can be given either the path to a file, or an open HDUList, as its first argument.
Applying a Headerlet Object
What gets done to a Headerlet object? The main purpose of a Headerlet object is to apply its WCS solution to the header of another FITS file. This can be done using the Headerlet.apply() method:
>>> hdrlet.apply(’some_other_image.fits’)
Or use the applyHeaderlet() convenience function that takes an existing headerlet file path or object as its first argument. The remaining arguments are the same as those used when calling Headerlet.apply(). As with createHeaderlet(), both of these can take a file path, or opened HDUList objects, as arguments.
Accessing the Headerlet Extension using PyFITS
When a headerlet is applied to an image, an additional headerlet containing that image’s original WCS solution is automatically created, and is appended to the file’s HDU list as a Headerlet HDU. However, this behavior can be disabled by setting the createheaderlet keyword argument to False in either Headerlet.apply() or applyHeaderlet().
When opening a file that contains Headerlet HDU extensions, it will normally appear in PyFits as follows:
>>> import pyfits >>> hdul = pyfits.open(’94f05bgq_flt_with_hlet.fits’) >>> hdul.info() Filename: j94f05bgq_flt_with_hlet.fits No. Name Type Cards Dimensions Format 0 PRIMARY PrimaryHDU 248 () int16 1 SCI ImageHDU 286 (4096, 2048) float32 2 ERR ImageHDU 76 (4096, 2048) float32 3 DQ ImageHDU 66 (4096, 2048) int16 4 SCI ImageHDU 282 (4096, 2048) float32 5 ERR ImageHDU 74 (4096, 2048) float32 6 DQ ImageHDU 66 (4096, 2048) int16 7 WCSCORR BinTableHDU 56 10R x 23C [40A,I,1A,D,D,D,D,D,D,D,D,24A, 24A,D,D,D,D,D,D,D,D,J,40A] 8 WCSDVARR ImageHDU 15 (65, 33) float32 9 WCSDVARR ImageHDU 15 (65, 33) float32 10 WCSDVARR ImageHDU 15 (65, 33) float32 11 WCSDVARR ImageHDU 15 (65, 33) float32 12 D2IMARR ImageHDU 12 (4096,) float32 13 HDRLET HeaderletHDU 13 14 HDRLET HeaderletHDU 13
The names of the separate headerlet extensions, reported as HeaderletHDUs, are both HDRLET, but its type shows up as HeaderletHDU when STWCS has been installed. The headers can be read, and although the data can be read, the user would have to know what to do with it, as the data has been converted to a tar file containing the headerlet FITS object.
If the stwcs.wcsutil.headerlet module is imported, though, PyFITS will recognize these extensions as special Headerlet HDUs and allow viewing of the data in the Headerlet. The summary of the FITS extensions for an image with a Headerlet HDU can be accessed using the following Python commands:
>>> import stwcs.wcsutil.headerlet >>> # Note that it is necessary to reopen the file >>> hdul = pyfits.open(’j94f05bgq_flt_with_hlet.fits’) >>> hdul.info() Filename: j94f05bgq_flt_with_hlet.fits No. Name Type Cards Dimensions Format 0 PRIMARY PrimaryHDU 248 () int16 1 SCI ImageHDU 286 (4096, 2048) float32 2 ERR ImageHDU 76 (4096, 2048) float32 3 DQ ImageHDU 66 (4096, 2048) int16 4 SCI ImageHDU 282 (4096, 2048) float32 5 ERR ImageHDU 74 (4096, 2048) float32 6 DQ ImageHDU 66 (4096, 2048) int16 7 WCSCORR BinTableHDU 56 10R x 23C [40A,I,1A D,D,D,D,D,D,D,D,24A, 24A,D,D,D,D,D,D,D,D,J,40A] 8 WCSDVARR ImageHDU 15 (65, 33) float32 9 WCSDVARR ImageHDU 15 (65, 33) float32 10 WCSDVARR ImageHDU 15 (65, 33) float32 11 WCSDVARR ImageHDU 15 (65, 33) float32 12 D2IMARR ImageHDU 12 (4096,) float32 13 HDRLET HeaderletHDU 13 14 HDRLET HeaderletHDU 13 >>> hdul[’HDRLET’, 1].header XTENSION= ’HDRLET ’ / Headerlet extension BITPIX = 8 / array data type NAXIS = 1 / number of array dimensions NAXIS1 = 102400 / Axis length PCOUNT = 0 / number of parameters GCOUNT = 1 / number of groups EXTNAME = ’HDRLET ’ / name of the headerlet extension HDRNAME = ’j94f05bgq_orig’ / Headerlet name DATE = ’2011-04-13T12:14:42’ / Date FITS file was generated SIPNAME = ’IDC_qbu1641sj’ / SIP distortion model name NPOLFILE= ’/grp/hst/acs/lucas/new-npl/qbu16424j_npl.fits’ / Non-polynomial corre D2IMFILE= ’/grp/hst/acs/lucas/new-npl/wfc_ref68col_d2i.fits’ / Column correction COMPRESS= F / Uses gzip compression
HeaderletHDU objects are similar to other HDU objects in PyFITS. However, they have a special .headerlet attribute that returns the actual FITS formatted headerlet contained in the HDU data as a Headerlet object as if the user were reading it directly from a standalone headerlet FITS file with a _hlet.fits filename. The .headerlet object can be read using the following Python syntax:
>>> hdrlet = hdul[’HDRLET’, 1].headerlet >>> hdrlet.info() Filename: (No file associated with this HDUList) No. Name Type Cards Dimensions Format 0 PRIMARY PrimaryHDU 12 () uint8 1 SIPWCS ImageHDU 111 () uint8 2 SIPWCS ImageHDU 110 () uint8 3 WCSDVARR ImageHDU 15 (65, 33) float32 4 WCSDVARR ImageHDU 15 (65, 33) float32 5 WCSDVARR ImageHDU 15 (65, 33) float32 6 WCSDVARR ImageHDU 15 (65, 33) float32 7 D2IMARR ImageHDU 12 (4096,) float32
This is useful for viewing the contents of the headerlets attached to a file without having to write out the Headerlet to its own separate FITS file, especially if this image was updated by someone else.
Other Headerlet Extension Functions
The presence of a HeaderletHDU (headerlet extension) in a science image’s FITS file indicates that at one point the science image’s WCS was updated using the information from that headerlet. In fact, many updates may have been applied to a science image’s WCS making it possible that multiple headerlet extensions may be present in the science image’s FITS file. The remaining functions provided by the stwcs.wcsutil.headerlet module allows a user to work with these multiple headerlet solutions to perform such operations as deleting extraneous headerlet extensions, write out the headerlet extension as a standalone FITS file, or even use a headerlet extension to update the WCS of the science image.
These additional capabilities can be broken down into two separate sets: use of the Headerlet object and use of headerlet FITS files. Once a Headerlet object has been created in memory, either from a .headerlet attribute of a HeaderletHDU as illustrated in the previous example or from reading in a headerlet FITS file, it can be used to perform several operations as defined by the following methods of the Headerlet class.
The functions provided by the headerlet module for working with headerlet FITS files covers all the same basic operations supported by the Headerlet class and its methods while using headerlet FITS files as direct inputs for each function. Unfortunately, providing full details on all the available operations and how to perform those operations go beyond the scope of this handbook at this time. Full details can be obtained from the documentation included with the software itself.
apply_as_alternate : function apply_as_alternate(self, fobj, attach=True, wcskey=None, wcsname=None)
This method allows a Headerlet object to be added to the list of alternate WCS solutions for the science data in the FITS file.
apply_as_primary : function apply_as_primary(self, fobj, attach=True, archive=True, force=False)
This method allows a Headerlet object to replace the primary(default) WCS of the science data in the FITS file.
attach_to_file : function attach_to_file(self, fobj, archive=False)
This method allows a Headerlet object to be added as a new HeaderletHDU extension in a science image's FITS file.
build_distname : function build_distname(self, dest)
This method creates the value for the DISTNAME keyword which gets used to identify what distortion model has been included in the headerlet.
hverify : function hverify(self)
This method verifies that the Headerlet object has all the necessary information in the PRIMARY header to be considered a valid headerlet.
info : function info(self, columns=None, pad=2, maxwidth=None, output=None, clobber=True, quiet=False)
A summary of the key distortion and WCS related keywords from the Headerlet will be reported in a table format.
summary : function summary(self, columns=None)
A summary of the key distortion and WCS related keywords from the Headerlet will be reported as a Python dictionary.
tofile : function tofile(self, fname, destim=None, hdrname=None, clobber=False)
A Headerlet object can be written out to it's own FITS file using this method.
verify_dest : function verify_dest(self, dest)
Use this method to verify whether or not this Headerlet can be applied to the destination science image based on the DESTNAME (or ROOTNAME) keywords.
verify_hdrname : function verify_hdrname(self, dest)
Use this method to verify whether or not the destination science image already has a headerlet applied to it with the same HDRNAME label.
verify_model : function verify_model(self, dest)
Use this method to verify whether or not this Headerlet can be applied to the destination science image by checking that the distortion model in the Headerlet matches the distortion model in the science image.
1 Retired instruments are available in Static Archives. Please refer to the MAST website for additional information. WFPC2 and NICMOS data can be reprocessed using DrizzlePac tasks.
2 For A_p_q, (p + q) ≤ A_ORDER; for B_p_q,, (p + q) ≤ B_ORDER
3 Residual or non-polynomial optical distortion corrections, contained in the NPOLFILE reference file and stored as image extensions of type WCSDVARR, are currently only implemented for ACS images. The column width corrections, from the D2IMFILE reference file and stored in image extension type D2IMARR, are only used for ACS/WFC images. Please check the DrizzlePac website for possible updates for other instruments. Note that WFPC2 images, currently in the static Archive, also requires the row width correction, but this is implemented directly by AstroDrizzle.
4 FITS-compliant files can be a multi-extension FITS file or an given extension to a multi-extension FITS file.